Bacterial Fluorescence meets the Kestrel™: How Image Processing is Helping Us See More in Less Time


Ever wonder how scientists can capture millions of tiny, glowing bacterial cells without losing their minds—or their weekends? Well, it’s not magic—it’s cutting-edge technology combined with some smart image processing techniques. Recently, Kristen Lok, a graduate student in the You lab at Duke University, worked on a project that harnesses the power of the Multi-Channel Automated Microscope (MCAM®), a high-tech tool that has drastically improved the speed and efficiency of imaging bacterial samples. And the best part? It’s making a huge difference in meeting the data demand for analyses in biological applications.
Faster Imaging for Faster Research
Biologists often need to capture images from multiple channels (like red and green fluorescence, and brightfield) to study how different proteins are expressed in bacteria. However, traditional imaging methods can take forever—imagine trying to image thousands of bacterial colonies in three different channels, with identical conditions across all samples. It would take days... or so it used to.
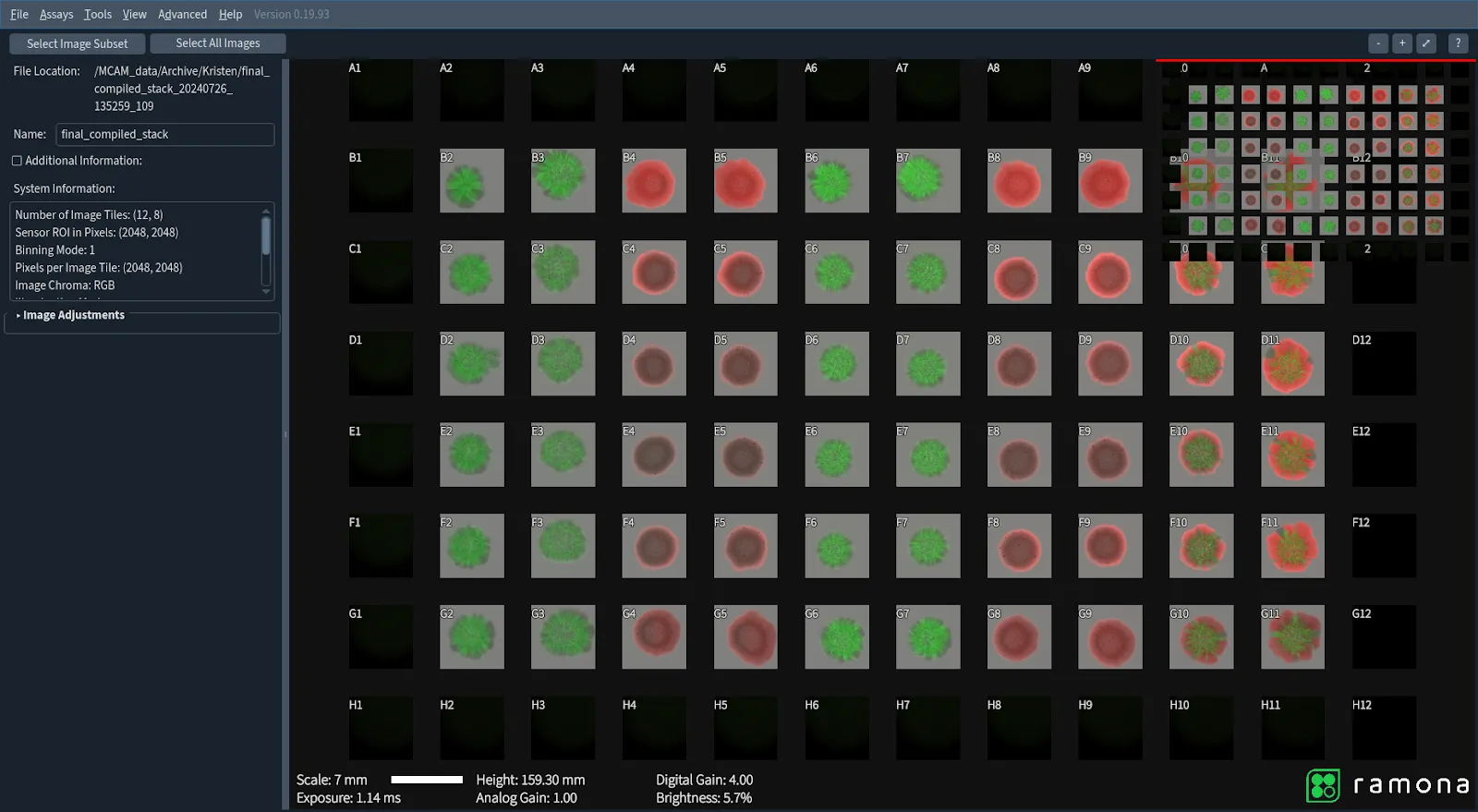
Enter the Kestrel powered by the MCAM technology, a powerhouse microscope that dramatically speeds up the process. By using this tool in another experiment, the team was able to cut the imaging time of 60 specimens contained within 34 plates down to just under two hours—a major efficiency boost (previously, image acquisitions for this many plates would take 11 hours spread over 2 days). The time saved allows researchers to focus on what really matters: analyzing the data and drawing meaningful conclusions from it.
The Challenge: Getting the Data Just Right
But speed isn’t everything. The next hurdle was ensuring the quality of the images. The data collected from the Kestrel needs to be combined in a way that preserves the real biological signals without any unintended enhancements. That’s especially tricky when you're dealing with different fluorescence channels like red and green, which can vary in brightness. Red fluorescence is typically dimmer than green, and if you're not careful, the green signal can overwhelm the red—or worse, the brightfield image.
While there are easy ways to stack these images using Python libraries like Pillow, they don’t give enough control over how the channels are combined. So, instead of just using the quick-and-easy solution, the team decided to take a more scientific approach to solve the problem.
The Solution: Calculating and Adjusting for Accuracy
The researchers started by calculating the correlation coefficient between the images from each channel. It allowed them to compare the images and to figure out how much adjustment each one needs. By separating each image into its three channels (brightfield, red, and green), they could apply the correlation coefficient to each channel individually. This adjustment ensures that the brightness and intensity of each channel are balanced accurately before they’re combined.
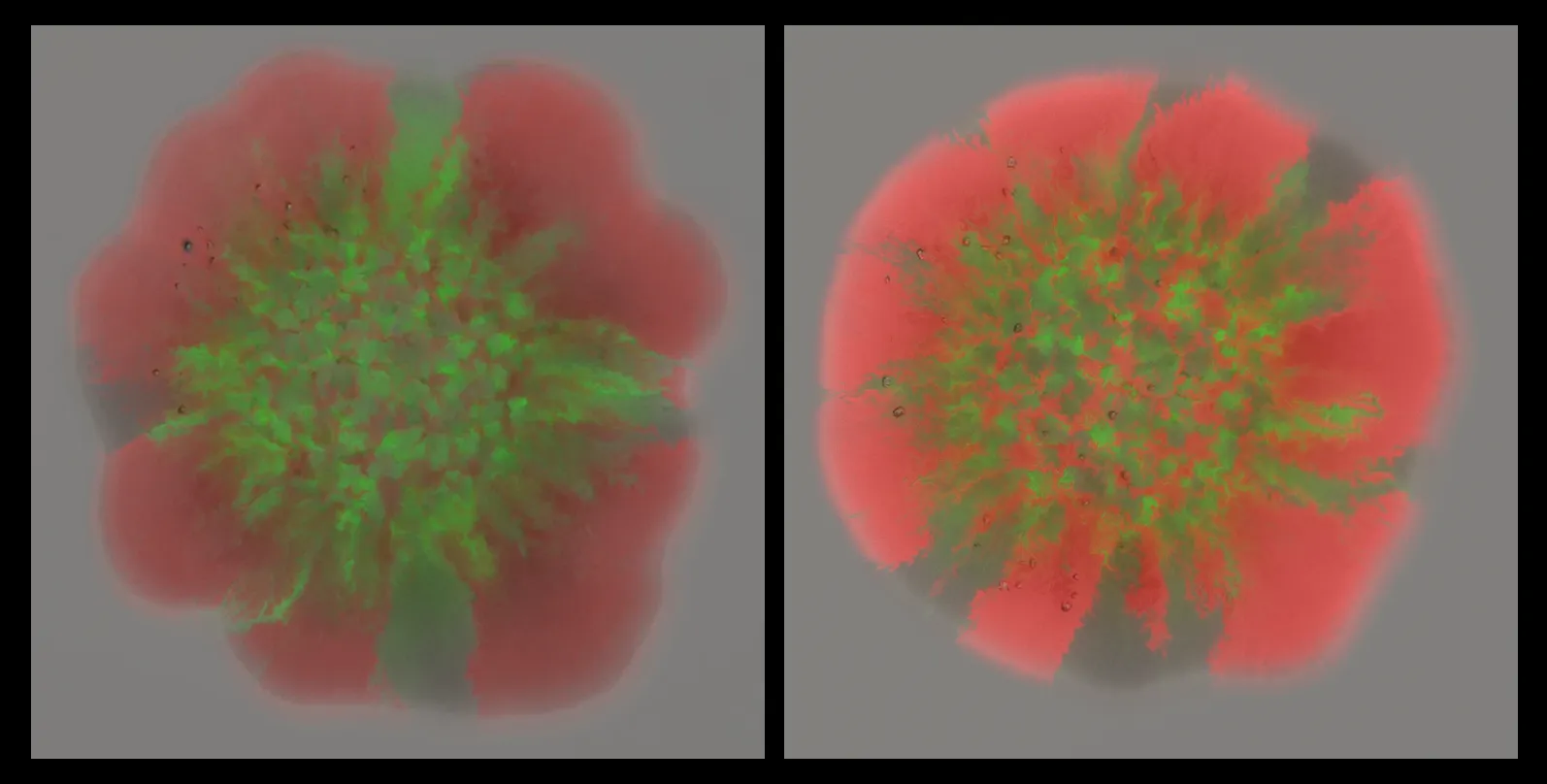
Finally, the team created a new image by summing the adjusted values from each channel, resulting in an overlay that accurately represented the real intensity of each one. And because this process was automated using a custom Python script, it took mere seconds to run for hundreds of images. In contrast, the old method could take an entire day to do the same thing.
.webp)
Why This Matters
The ability to quickly and accurately process these images is a game-changer for the research community. Not only does it save time, but it also helps researchers better learn from bacteria using fluorescence information. This is especially important for studying the morphology and development of bacterial colonies with unique gene expression and under unique growth conditions.
In the end, this combination of advanced microscopy and smart image processing is enabling scientists to push the boundaries of biological research, opening the door to faster discoveries and more accurate insights. And all it took was a new ultrafast microscope, a little bit of clever coding and a whole lot of bacteria.

Ever wonder how scientists can capture millions of tiny, glowing bacterial cells without losing their minds—or their weekends? Well, it’s not magic—it’s cutting-edge technology combined with some smart image processing techniques. Recently, Kristen Lok, a graduate student in the You lab at Duke University, worked on a project that harnesses the power of the Multi-Channel Automated Microscope (MCAM®), a high-tech tool that has drastically improved the speed and efficiency of imaging bacterial samples. And the best part? It’s making a huge difference in meeting the data demand for analyses in biological applications.
Faster Imaging for Faster Research
Biologists often need to capture images from multiple channels (like red and green fluorescence, and brightfield) to study how different proteins are expressed in bacteria. However, traditional imaging methods can take forever—imagine trying to image thousands of bacterial colonies in three different channels, with identical conditions across all samples. It would take days... or so it used to.

Enter the Kestrel powered by the MCAM technology, a powerhouse microscope that dramatically speeds up the process. By using this tool in another experiment, the team was able to cut the imaging time of 60 specimens contained within 34 plates down to just under two hours—a major efficiency boost (previously, image acquisitions for this many plates would take 11 hours spread over 2 days). The time saved allows researchers to focus on what really matters: analyzing the data and drawing meaningful conclusions from it.
The Challenge: Getting the Data Just Right
But speed isn’t everything. The next hurdle was ensuring the quality of the images. The data collected from the Kestrel needs to be combined in a way that preserves the real biological signals without any unintended enhancements. That’s especially tricky when you're dealing with different fluorescence channels like red and green, which can vary in brightness. Red fluorescence is typically dimmer than green, and if you're not careful, the green signal can overwhelm the red—or worse, the brightfield image.
While there are easy ways to stack these images using Python libraries like Pillow, they don’t give enough control over how the channels are combined. So, instead of just using the quick-and-easy solution, the team decided to take a more scientific approach to solve the problem.
The Solution: Calculating and Adjusting for Accuracy
The researchers started by calculating the correlation coefficient between the images from each channel. It allowed them to compare the images and to figure out how much adjustment each one needs. By separating each image into its three channels (brightfield, red, and green), they could apply the correlation coefficient to each channel individually. This adjustment ensures that the brightness and intensity of each channel are balanced accurately before they’re combined.

Finally, the team created a new image by summing the adjusted values from each channel, resulting in an overlay that accurately represented the real intensity of each one. And because this process was automated using a custom Python script, it took mere seconds to run for hundreds of images. In contrast, the old method could take an entire day to do the same thing.
.webp)
Why This Matters
The ability to quickly and accurately process these images is a game-changer for the research community. Not only does it save time, but it also helps researchers better learn from bacteria using fluorescence information. This is especially important for studying the morphology and development of bacterial colonies with unique gene expression and under unique growth conditions.
In the end, this combination of advanced microscopy and smart image processing is enabling scientists to push the boundaries of biological research, opening the door to faster discoveries and more accurate insights. And all it took was a new ultrafast microscope, a little bit of clever coding and a whole lot of bacteria.
.png)
.webp)