White Paper: Live Cardiac Organoid Fluorescence Video Capture and Analysis with Multi-Camera Array Microscopes

Introduction
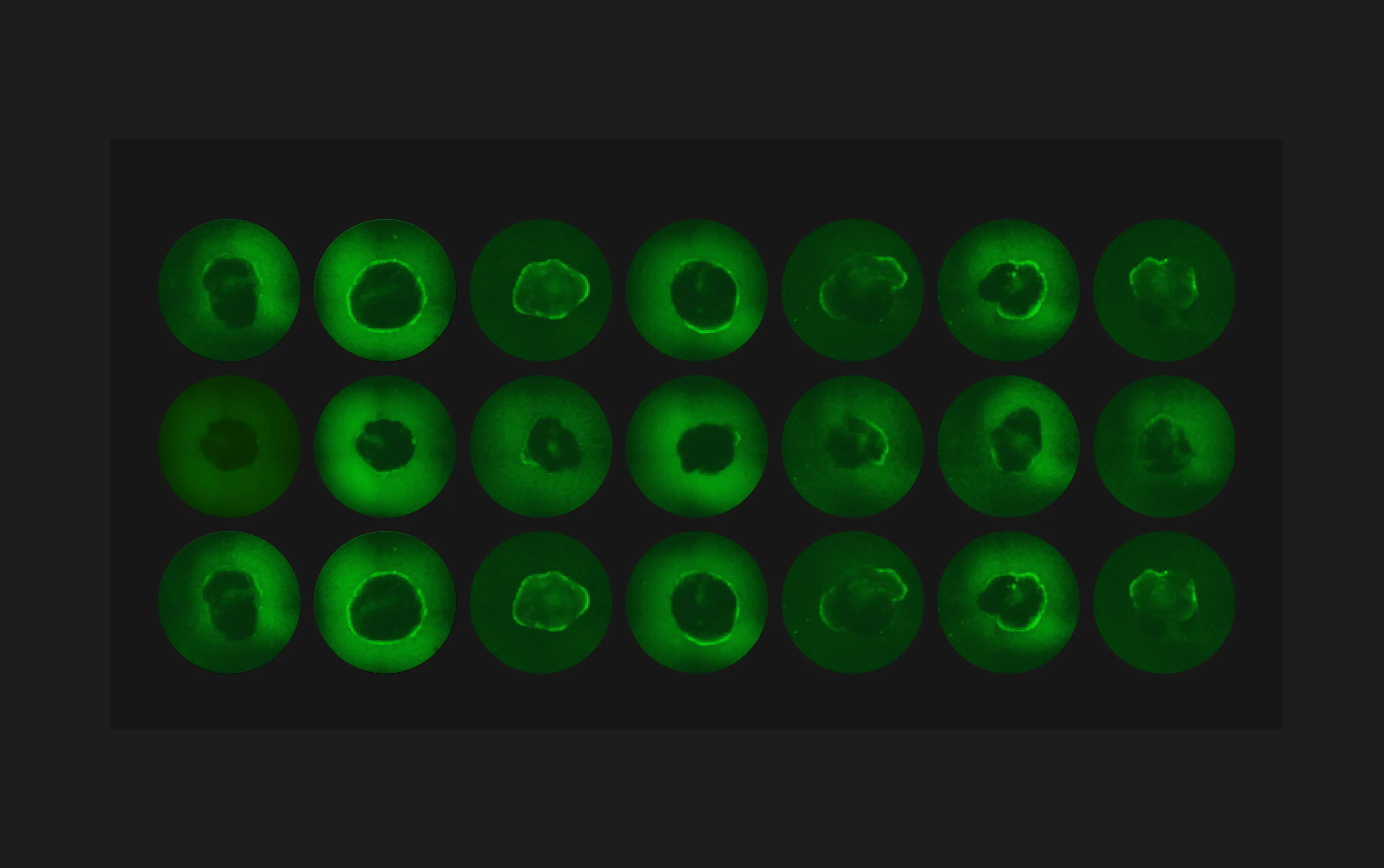
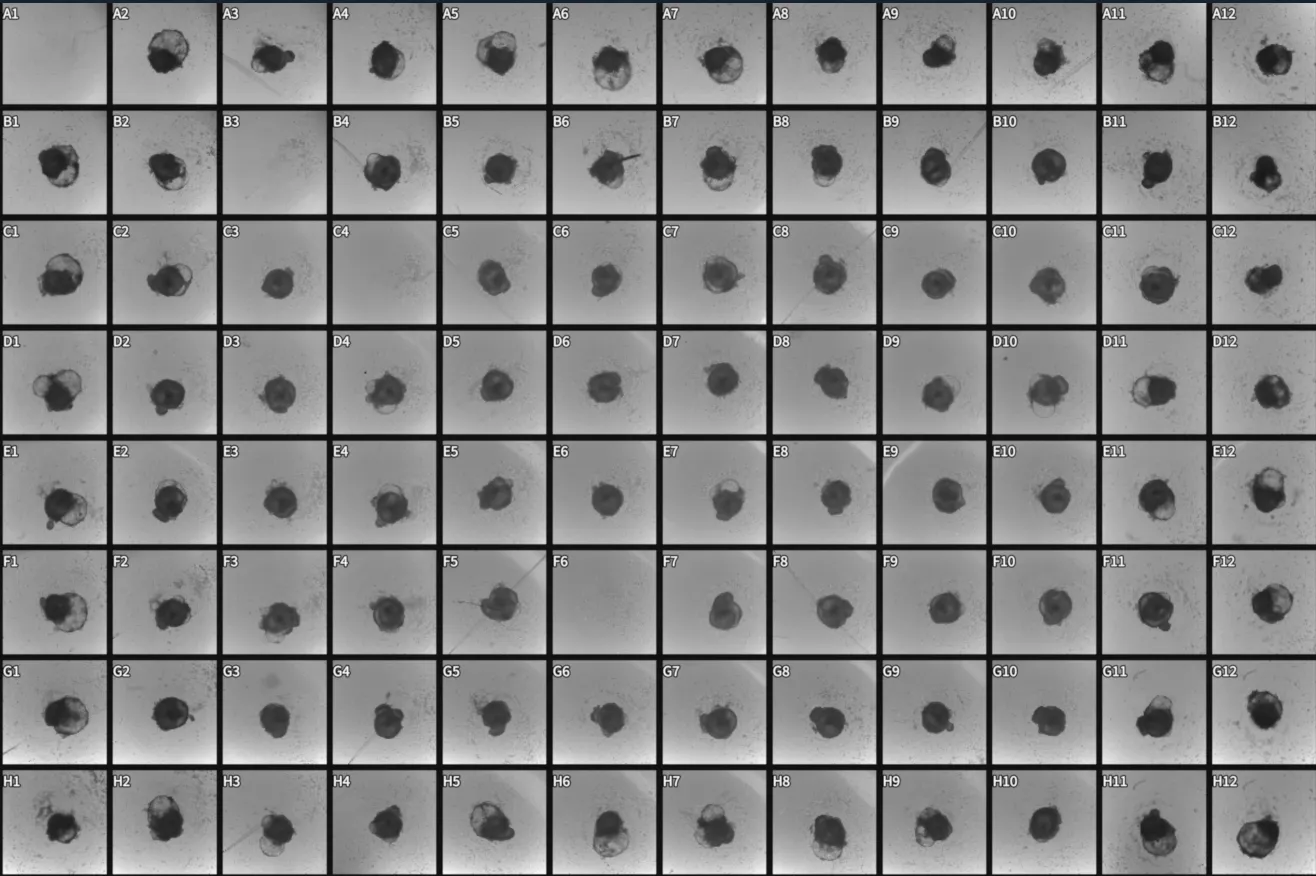
Recent advances in pluripotent stem cell-derived human cardiac organoids have provided researchers with a valuable tool in studies with cardiac diseases. However, large-scale functional screening of cardiac organoid beating and calcium firing are limited by the imaging speed of what a conventional microscope can achieve across multiple wells, especially those that require temporal precision. Furthermore, imaging cardiac organoids individually inadvertently introduce confounding temporal factors, thus limiting time-sensitive comparative studies across wells. To overcome the limitation, we utilize Ramona’sVireo™ system, which leverages our proprietary Multi-Camera Array Microscope (MCAM®) technology. The system includes 24 objectives organized in a compact array that allows video-recordings of cardiac organoids in 24 wells simultaneously (Figure 1).
Using the Vireo, we recorded cardiac organoid contractions and calcium flux in a green fluorescence channel (440nm GFP) with a frame rate of 45 frames per second (FPS) at a 1.17μm/pixel resolution, binx2 (effectively 2.34μm/pixel) for 10 seconds per groupings, with a total of 90 seconds including stage movement and video encoding for all 96-wells. We then processed the videos using the MCAM Software's Regions of Interest tool followed by comprehensive quantitative analyses to demonstrate three key technological advantages: (1) simultaneous multi-well recording capability, (2) high-quality functional fluorescence imaging, and (3) seamless data compatibility with downstream analysis software.
Our comprehensive validation demonstrates the system's versatility across multiple analysis paradigms. The high-resolution fluorescence imaging capabilities capture subtle spatial heterogeneities within individual organoids, while the rapid acquisition rate ensures temporal fidelity for kinetic analyses. Most importantly, the system's commitment to standard data formats ensure that researchers can seamlessly integrate Vireo-acquired data into existing computational pipelines, whether using Python-based analysis frameworks, ImageJ workflows, or specialized cardiac analysis software.


Methods and Results
Experimental Setup and Video Capturing
Cardiac Organoid Culture: Human iPSC-derived cardiac organoids were cultured using established protocols and maintained in 96-well plates. Fluo-4-AM was added to the wells to enable real-time calcium flux monitoring during spontaneous beating activity.
Simultaneous Video Recording Protocol: The Vireo system was configured for simultaneous 24-well imaging using the following parameters:
- Frame rate: 45 frames per second (FPS)
- Spatial resolution: 1.17 μm/pixel, binx2 (effectively 2.34μm/pixel)
- Recording duration: 10 seconds per acquisition
- Illumination mode: Fluorescence Ex 400nm — GFP FITC
- Synchronization: Simultaneous acquisition across all 24 cameras
- Data export: Standard video formats (MP4, TIFF stacks) with embedded metadata
Computational Analysis with Python
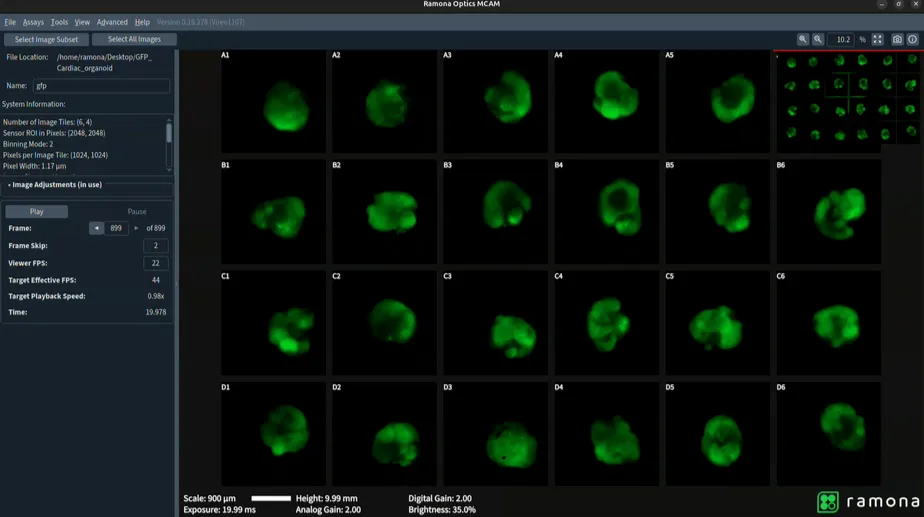
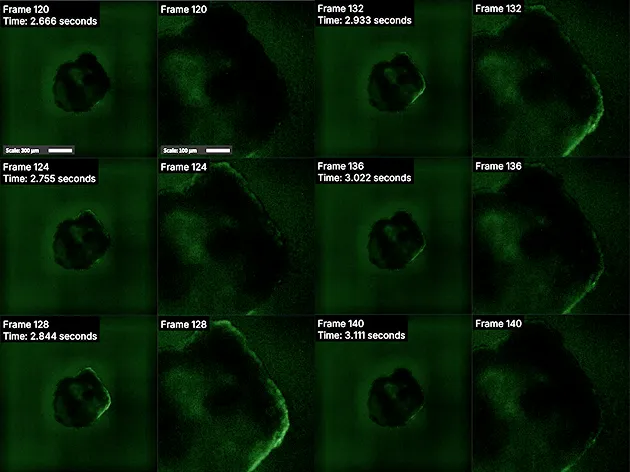
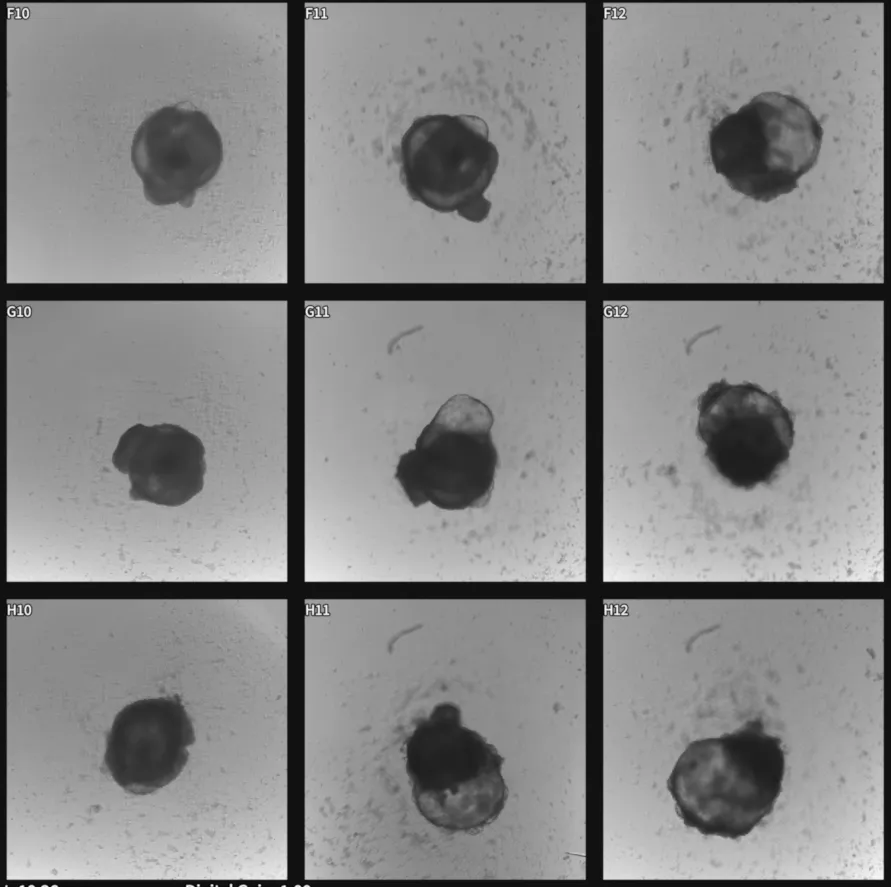
Video recordings captured using the Vireo were opened in MCAM viewer software where regions of interest were selected using the Region of Interest Tool (Figure 3). The subsequent videos containing only selected regions (Figure 4) were then analyzed through a Python-based computational pipeline implemented within a Jupyter Notebook environment that allows users to easily load MP4/Tiff video files directly from an MCAM acquisition to calculate mean fluorescence per frame, normalize to ΔF/F₀ using 10th-percentile baseline, and plot fluorescence dynamics over time. The Jupyter Notebook uses standard Python libraries. (imageio, numpy, matplotlib).
Raw MP4 files output by the MCAM viewer software were imported using the imageio library. Mean fluorescence intensities were computed for each frame, quantifying dynamic signal variations over time. Temporal synchronization was ensured by extracting the frame rate (FPS) from the video metadata, providing a precise temporal axis. Fluorescence changes were normalized to a baseline defined by the 10th percentile intensity, a measure that prevents inaccuracies when initial frames lack a clear fluorescence signal, improving reliability over conventional first-frame normalization. Plotting with Matplotlib demonstrated fluorescence intensity dynamics, highlighting cardiac contractility and calcium signaling events, and supporting meaningful comparative analyses across experimental conditions (Figure 6).

.webp)
Conclusions
System Performance Validation
The Vireo system successfully recorded synchronized calcium dynamics across 24 wells simultaneously. The complete 96-well plate acquisition required only 90 seconds total, representing a significant improvement over sequential imaging approaches that typically require 30-45 minutes for equivalent data collection.
Quantitative Calcium Signaling Analysis
The computational analysis pipeline successfully quantified dynamic fluorescence variations indicative of cardiac contractility and calcium signaling in human cardiac organoids. Analysis revealed clear, periodic peaks in fluorescence intensity corresponding to calcium transients during spontaneous contractions. Normalization to the robust baseline facilitated accurate comparisons across wells and conditions, minimizing variability due to baseline fluctuations. Notably, the temporal precision enabled by the synchronized multi-camera recording captured subtle differences in contractile kinetics and calcium signaling dynamics, allowing for robust statistical evaluation of cardiac organoid performance across diverse experimental treatments. These findings demonstrate the effectiveness of the computational pipeline in extracting meaningful physiological parameters from high-throughput imaging data, validating its utility in large-scale functional screening assays.

White Paper: Live Cardiac Organoid Fluorescence Video Capture and Analysis with Multi-Camera Array Microscopes
Introduction
Recent advances in pluripotent stem cell-derived human cardiac organoids have provided researchers with a valuable tool in studies with cardiac diseases. However, large-scale functional screening of cardiac organoid beating and calcium firing are limited by the imaging speed of what a conventional microscope can achieve across multiple wells, especially those that require temporal precision. Furthermore, imaging cardiac organoids individually inadvertently introduce confounding temporal factors, thus limiting time-sensitive comparative studies across wells. To overcome the limitation, we utilize Ramona’sVireo™ system, which leverages our proprietary Multi-Camera Array Microscope (MCAM®) technology. The system includes 24 objectives organized in a compact array that allows video-recordings of cardiac organoids in 24 wells simultaneously (Figure 1).
Using the Vireo, we recorded cardiac organoid contractions and calcium flux in a green fluorescence channel (440nm GFP) with a frame rate of 45 frames per second (FPS) at a 1.17μm/pixel resolution, binx2 (effectively 2.34μm/pixel) for 10 seconds per groupings, with a total of 90 seconds including stage movement and video encoding for all 96-wells. We then processed the videos using the MCAM Software's Regions of Interest tool followed by comprehensive quantitative analyses to demonstrate three key technological advantages: (1) simultaneous multi-well recording capability, (2) high-quality functional fluorescence imaging, and (3) seamless data compatibility with downstream analysis software.
Our comprehensive validation demonstrates the system's versatility across multiple analysis paradigms. The high-resolution fluorescence imaging capabilities capture subtle spatial heterogeneities within individual organoids, while the rapid acquisition rate ensures temporal fidelity for kinetic analyses. Most importantly, the system's commitment to standard data formats ensure that researchers can seamlessly integrate Vireo-acquired data into existing computational pipelines, whether using Python-based analysis frameworks, ImageJ workflows, or specialized cardiac analysis software.



Methods and Results
Experimental Setup and Video Capturing
Cardiac Organoid Culture: Human iPSC-derived cardiac organoids were cultured using established protocols and maintained in 96-well plates. Fluo-4-AM was added to the wells to enable real-time calcium flux monitoring during spontaneous beating activity.
Simultaneous Video Recording Protocol: The Vireo system was configured for simultaneous 24-well imaging using the following parameters:
- Frame rate: 45 frames per second (FPS)
- Spatial resolution: 1.17 μm/pixel, binx2 (effectively 2.34μm/pixel)
- Recording duration: 10 seconds per acquisition
- Illumination mode: Fluorescence Ex 400nm — GFP FITC
- Synchronization: Simultaneous acquisition across all 24 cameras
- Data export: Standard video formats (MP4, TIFF stacks) with embedded metadata
Computational Analysis with Python
Video recordings captured using the Vireo were opened in MCAM viewer software where regions of interest were selected using the Region of Interest Tool (Figure 3). The subsequent videos containing only selected regions (Figure 4) were then analyzed through a Python-based computational pipeline implemented within a Jupyter Notebook environment that allows users to easily load MP4/Tiff video files directly from an MCAM acquisition to calculate mean fluorescence per frame, normalize to ΔF/F₀ using 10th-percentile baseline, and plot fluorescence dynamics over time. The Jupyter Notebook uses standard Python libraries. (imageio, numpy, matplotlib).
Raw MP4 files output by the MCAM viewer software were imported using the imageio library. Mean fluorescence intensities were computed for each frame, quantifying dynamic signal variations over time. Temporal synchronization was ensured by extracting the frame rate (FPS) from the video metadata, providing a precise temporal axis. Fluorescence changes were normalized to a baseline defined by the 10th percentile intensity, a measure that prevents inaccuracies when initial frames lack a clear fluorescence signal, improving reliability over conventional first-frame normalization. Plotting with Matplotlib demonstrated fluorescence intensity dynamics, highlighting cardiac contractility and calcium signaling events, and supporting meaningful comparative analyses across experimental conditions (Figure 6).




.webp)
Conclusions
System Performance Validation
The Vireo system successfully recorded synchronized calcium dynamics across 24 wells simultaneously. The complete 96-well plate acquisition required only 90 seconds total, representing a significant improvement over sequential imaging approaches that typically require 30-45 minutes for equivalent data collection.
Quantitative Calcium Signaling Analysis
The computational analysis pipeline successfully quantified dynamic fluorescence variations indicative of cardiac contractility and calcium signaling in human cardiac organoids. Analysis revealed clear, periodic peaks in fluorescence intensity corresponding to calcium transients during spontaneous contractions. Normalization to the robust baseline facilitated accurate comparisons across wells and conditions, minimizing variability due to baseline fluctuations. Notably, the temporal precision enabled by the synchronized multi-camera recording captured subtle differences in contractile kinetics and calcium signaling dynamics, allowing for robust statistical evaluation of cardiac organoid performance across diverse experimental treatments. These findings demonstrate the effectiveness of the computational pipeline in extracting meaningful physiological parameters from high-throughput imaging data, validating its utility in large-scale functional screening assays.
